The asymptor R package allows you to estimate the lower and upper bound of asymptomatic cases in an epidemic using the capture/recapture methods from Böhning et al. (2020) and Rocchetti et al. (2020).
Please note there is currently some discussion about the validity of the methods implemented in this package. You should read carefully the original articles, alongside this answer from Li et al. (2022) before using this package in your project.
Installation
You can install the stable version of this package from CRAN:
install.packages("asymptor")or the development version from GitHub, via my r-universe:
install.packages("asymptor", repos = "https://bisaloo.r-universe.dev")Example
Let’s start by loading some example data from the COVID-19 epidemic in Italy:
d <- readRDS(system.file("extdata", "covid19_italy.rds", package = "asymptor"))
head(d)
#> date new_cases new_deaths
#> 1 2020-01-02 0 0
#> 2 2020-01-03 0 0
#> 3 2020-01-04 0 0
#> 4 2020-01-05 0 0
#> 5 2020-01-06 0 0
#> 6 2020-01-07 0 0We can estimate the lower and upper bound of asymptomatic cases with:
library(asymptor)
estimate_asympto(d$date, d$new_cases, d$new_deaths)Or, with a tidyverse-compatible syntax:
library(dplyr)
d %>%
mutate(asympto_cases = estimate_asympto(date, new_cases, new_deaths))Please refer to the vignette for a detailed example using the COVID-19 data from Italy.
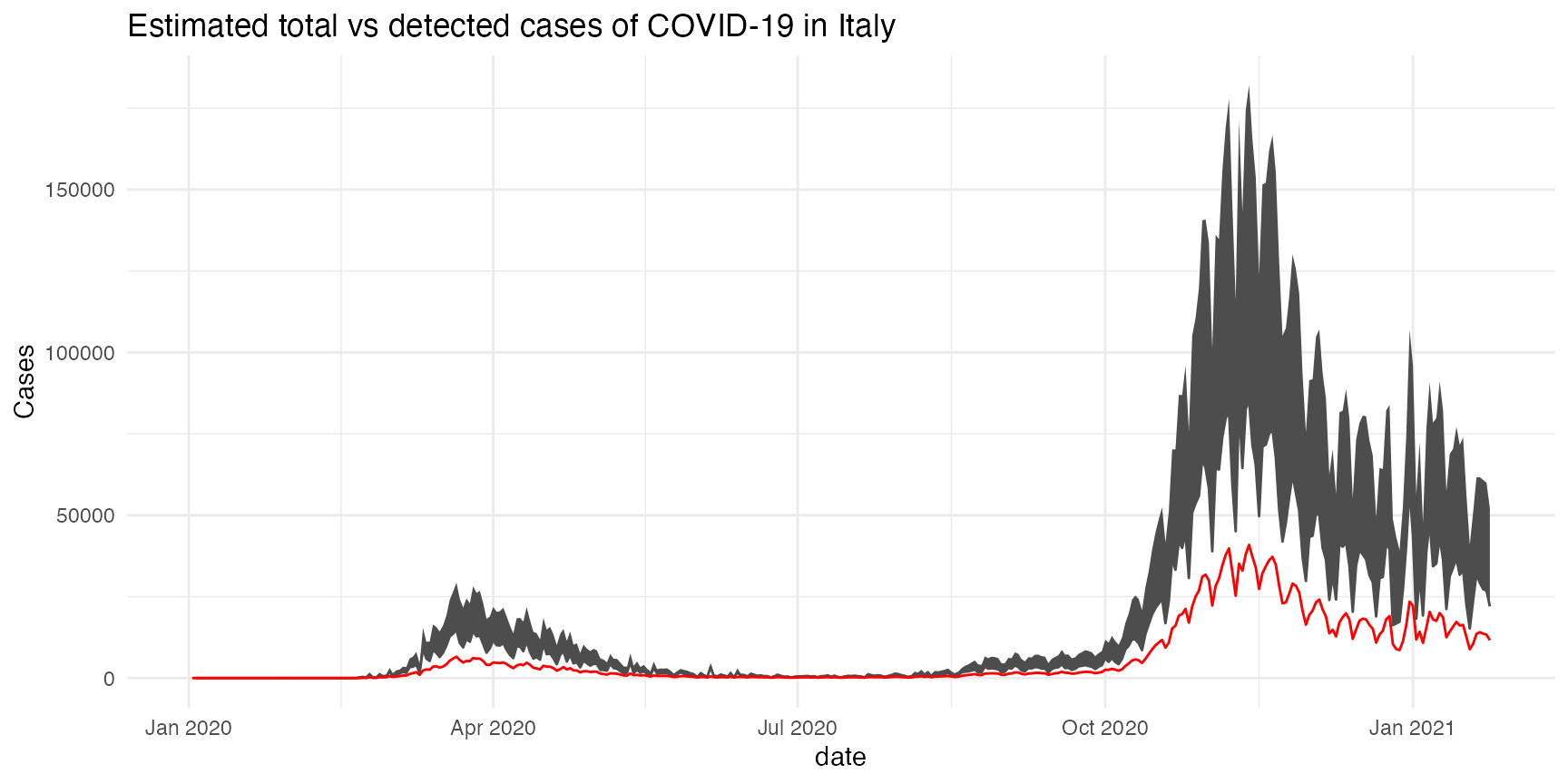
example_figure

